AI-driven design and automation, built for discovery

Data-efficient, physics-informed AI
We integrate physics principles in our AI architectures, creating more reliable and accurate models to venture deeper in chemical space.
Protein-ligand structure prediction3D physics-based equivariant generative diffusion
AI-accelerated quantum chemistryGraph neural network architecture based on quantum features
Multi-parameter lead selection
Massively pre-trained graph neural network, deployed across dozens of drug properties for lead optimization, and supported by automated training, uncertainty quantification and explainability
State-of-the-art data efficiency
Generative molecular design
Suite of generative technologies, fully aware of the chemical space efficiently accessible through our high-throughput chemistry platform
Optimally exploring chemical space, while ensuring rapid and successful execution on our platform
New molecular designs to new biological data each week
New molecular designs to new biological data each week
Parallel testing across alternative profiles based on platform-generated biological insights
De novo protein-ligand structure and cryptic pocket prediction
Nanoscale synthesis of thousands of unique compounds per week
Generating biochemical, metabolic, and cellular data
Automated data pipelines and model retraining, every week
Creating the right molecule for the right profile
By identifying diverse chemical matter with unique profiles, we arrive at highly differentiated development candidates selected from multiple strong lead series.
Publications
Learn more about the technology that drives our platform
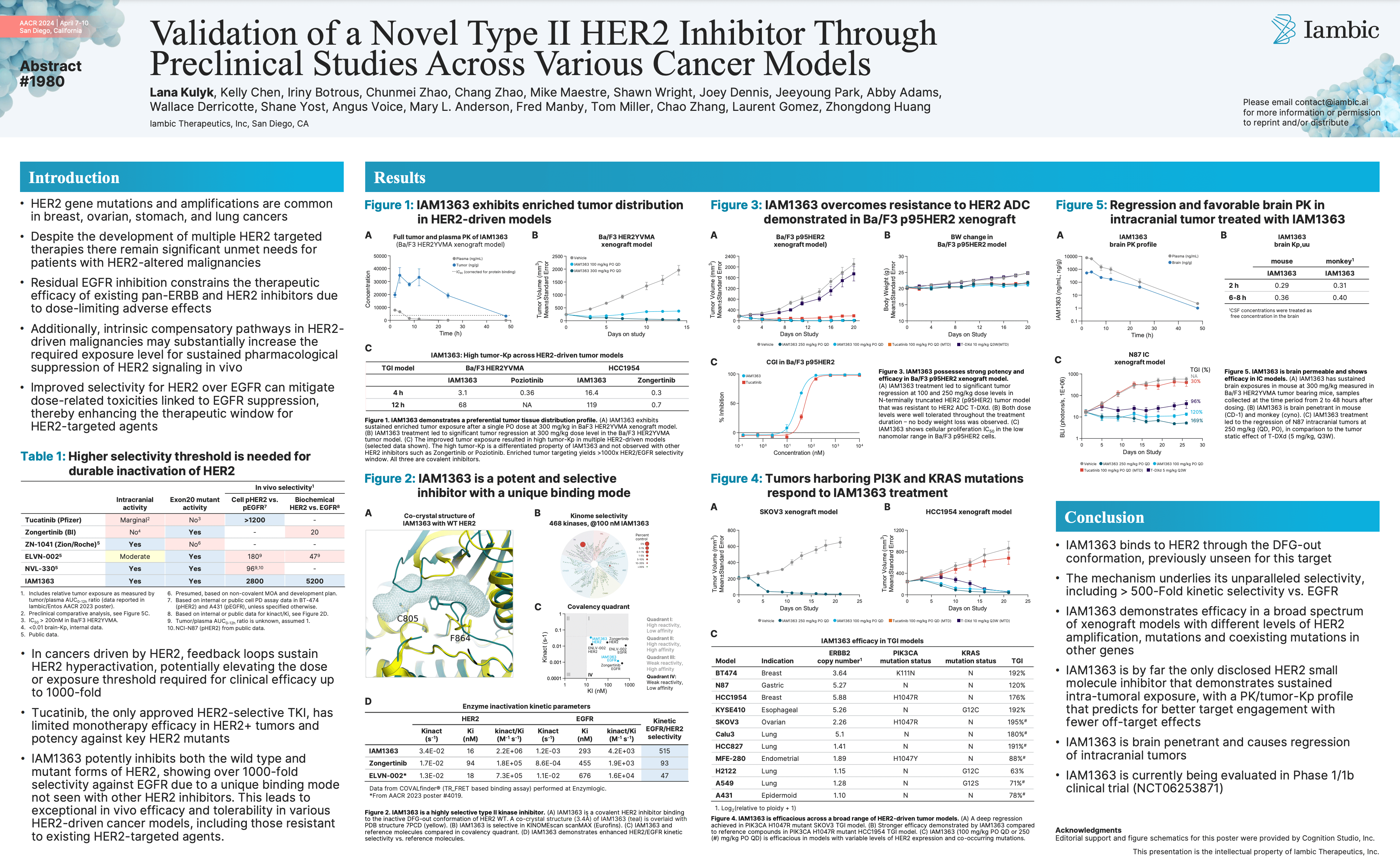
Validation of a Novel Type II HER2 Inhibitor Through Preclinical Studies Across Various Cancer Models (AACR Poster)

Validation of a Novel Type II HER2 Inhibitor Through Preclinical Studies Across Various Cancer Models (AACR Poster)

Iambic Therapeutics Announces New Research with Cover Article in Nature Machine Intelligence Demonstrating the Capabilities of its Generative AI NeuralPLexer Technology to Predict Protein-Ligand Complex Structures


.jpg)
.jpg)